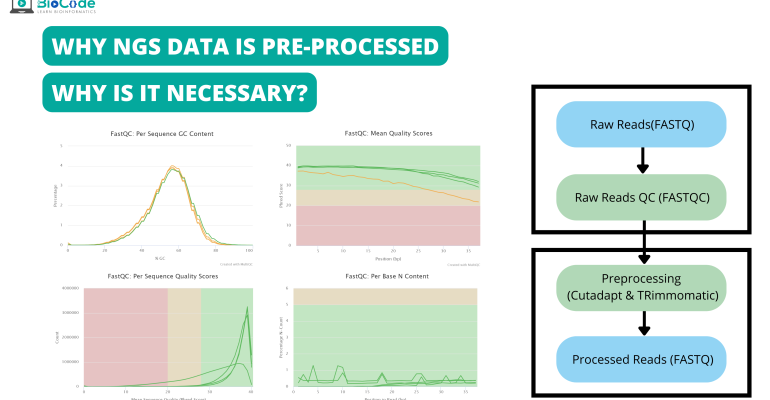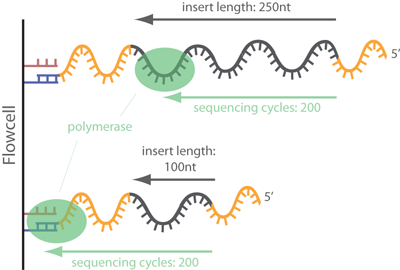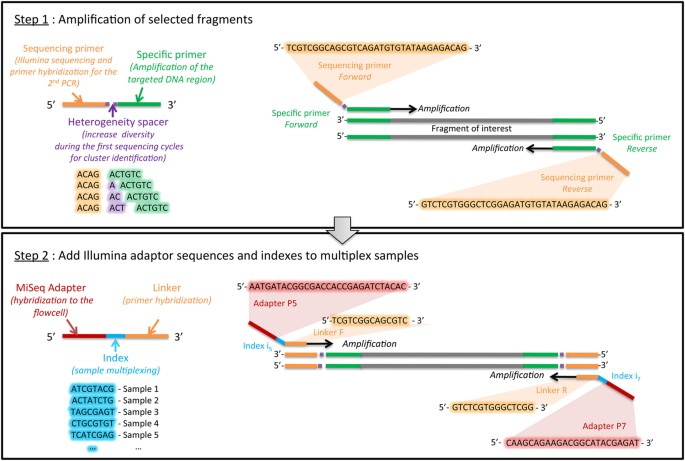
High-throughput sequencing of multiple amplicons for barcoding and integrative taxonomy | Scientific Reports
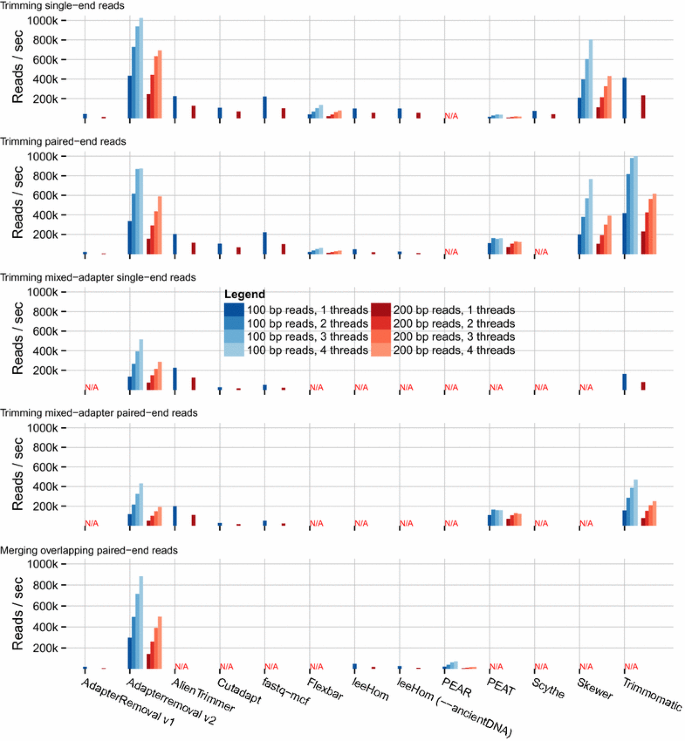
AdapterRemoval v2: rapid adapter trimming, identification, and read merging | BMC Research Notes | Full Text

Kraken: A set of tools for quality control and analysis of high-throughput sequence data☆ | Semantic Scholar
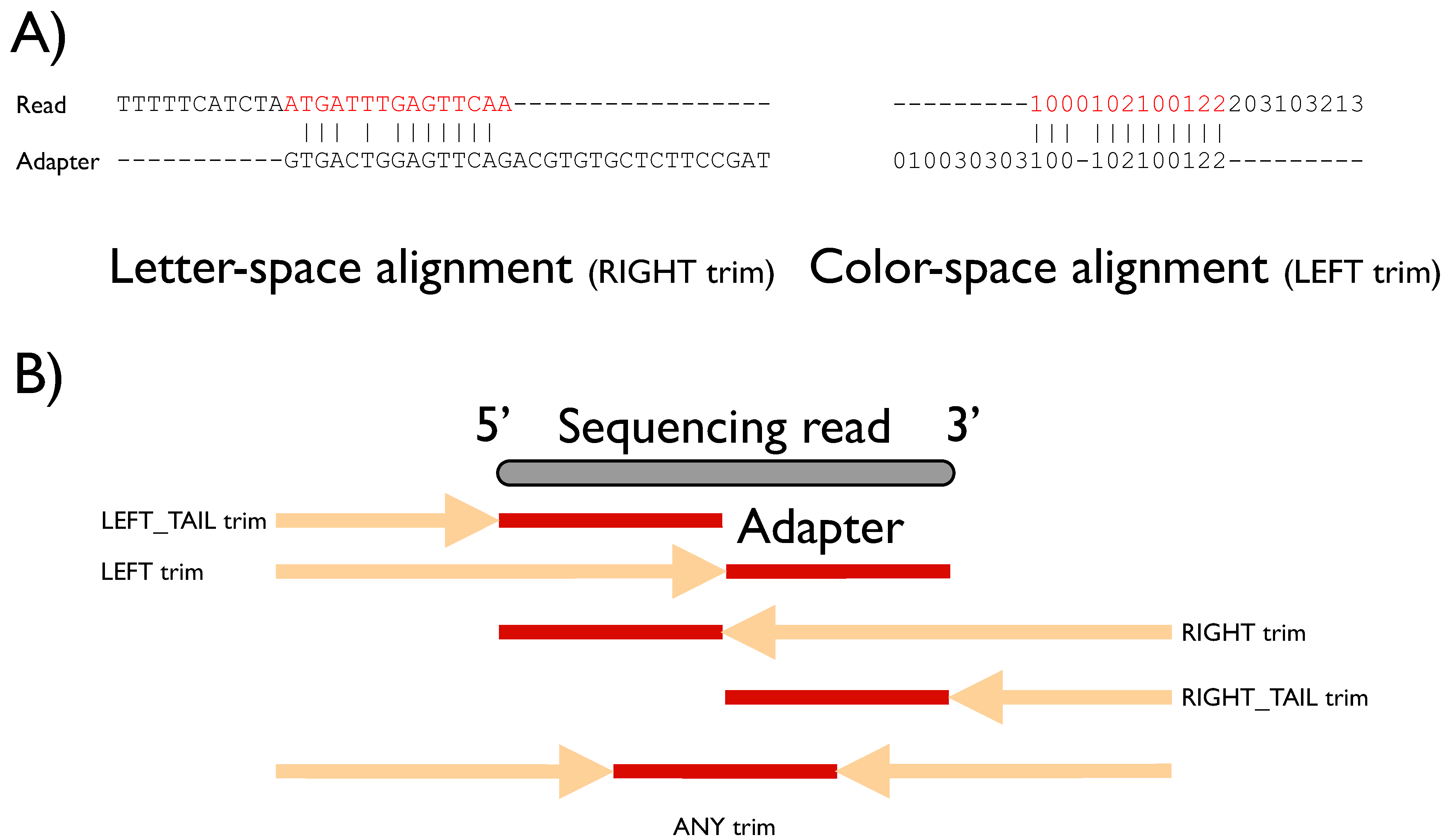
Biology | Free Full-Text | FLEXBAR—Flexible Barcode and Adapter Processing for Next-Generation Sequencing Platforms
Comparison of tools cutting primer sequences from reads with tolerance... | Download Scientific Diagram

Assessing the utility of long-read nanopore sequencing for rapid and efficient characterization of mobile element insertions - Laboratory Investigation
Cutadapt remoYes adapter sequences from high-throughput sequencing reads Introduction Implementation Features
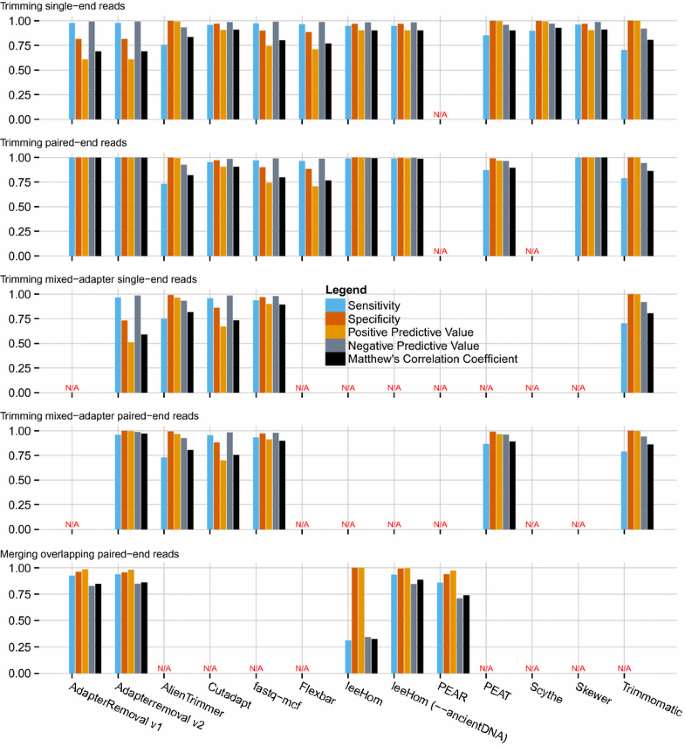
![PDF] leeHom: adaptor trimming and merging for Illumina sequencing reads | Semantic Scholar PDF] leeHom: adaptor trimming and merging for Illumina sequencing reads | Semantic Scholar](https://d3i71xaburhd42.cloudfront.net/247973613dfa3c42eb437ec9f5269202595f96af/2-Figure1-1.png)